The adsorption of proteins to surfaces, especially lipid layers, plays a critical role in biochemical processes within living systems. These include membrane signaling and recognition, toxin detection, and mitigating the effects of toxin/biowarfare agents. Specific issues to be addressed are: protein orientation and surface density, monolayer versus multilayer adsorption, specific versus non-specific interactions, and lateral order and conformational changes upon adsorption including denaturization of protein (Refer to reference 1).
In this work neutron reflection is combined with grazing incidence x-ray diffraction (G I X D) to study the interactions of the protein myoglobin with Langmuir monolayers of synthetic lipids. Neutron reflection is used to determine the adsorbed amount and to obtain information regarding the orientation and conformation of the adsorbed myoglobin (Refer to reference 2). G I X D is used to study the response of the lipid layer that occurs upon protein binding.
For lipid layers we have used the synthetic lipid distearyl imino-diacetate (D S I D A) that contains receptors for the metal ion copper 2 +. The use of metal ion coordination to target the adsorption of proteins to lipid membranes has been studied extensively. This method uses specific coordination interactions between copper 2 + and naturally occurring histidine units in myoglobin.
A Langmuir monolayer of 100 % D S I D A was spread onto the surface of buffered deuterated water or plain water (Figure 1) and then compressed to a surface pressure of approximately 40 milli newtons per meter where it forms a solid condensed phase layer (see G I X D data below). A dilute solution of copper chloride was injected under the condensed D S I D A monolayer. The copper 2 + ions chelate at the imino-diacetate site as indicated in Figure 1.
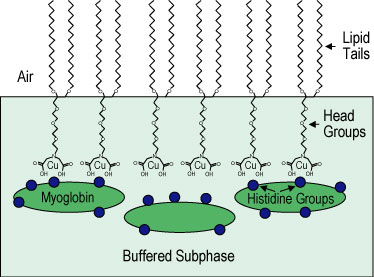
| Figure 1. Schematic of histidine groups on myoglobin adsorbing to distearyl imino-diacetate (D S I D A) molecules at chelated copper 2 + ion sites. |
Neutron reflectivity data for the condensed lipid layer with added copper 2 + ions are shown in Figure 2 for both the plain water and deuterated water buffered subphases. Myoglobin was then injected into the subphase and neutron reflectivity data were taken repeatedly to monitor the time dependence of protein adsorption. The adsorbed protein layers reached quasi-equilibrium at approximately 14 h after adding myoglobin into the solution. Only final sets of reflectivity data with adsorbed protein (for both plain water and deuterated water subphases) are shown in Figure 2. The very large change in the reflectivity after protein injection indicates a significant adsorption of protein under the lipid layer.
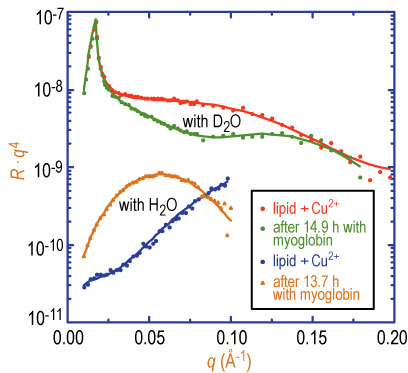
| Figure 2. Neutron reflectivity for 100 % D S I D A on buffered deuterated water and plain water subphases with and without myoglobin in the subphase. Solid lines are fits to the data as described in the text. |
Solid lines in Figure 2 are fits to the reflectivity data from which the segment concentration profile of protein can be obtained. Simultaneous fits to both sets of data show that the dimension of the quasi-equilibrium adsorbed myoglobin layer is 36 angstroms, plus or minus 2 angstroms, and that the segment volume fraction of protein is approximately 50 %. Unit cell dimensions of myoglobin from its crystal structure are A = 64.84 angstroms, B = 30.98 angstroms, and C = 34.92 angstroms. Thus, the protein in all likelihood is bound to the lipid layer in its native state, i.e., not denatured.
If no copper chloride is added to the subphase, then no change in the neutron reflectivity signal is observed upon injection of myoglobin. Alternatively, if we inject a strong copper scavenging agent (E D T A) in the subphase after the protein adsorption, the reflectivity reverts back to that of the pure condensed lipid layer. This clearly shows that we are observing very specific binding of the protein to the chelated copper 2 + sites on D S I D A..
Important insight has been revealed in complementary G I X D data from the D S I D A system. Figure 3 A, shows the surface diffraction peak from the tails of a condensed D S I D A monolayer at an in-plane wave vector Q, sub x y, = 1.50 inverse angstroms. This corresponds to a hexagonal packed lipid layer with a nearest neighbor spacing of 4.21 angstroms. Insertion of copper 2 + ions into the layer does not seem to affect the 2-D crystalline order of the lipid layer except for a slight change in lattice spacing. When myoglobin is added to the subphase at a constant surface pressure of 40 milli newtons per meter, the diffraction peak arising from the hexagonal packing of the D S I D A tails (at Q sub x y = 1.50 inverse angstroms) is drastically reduced at short times (Figure 3 b). This indicates a large perturbation in the 2-D crystallinity of the lipid layer upon insertion of protein. The surface diffraction peak, however, reappears after a few hours (Figure 3 b). The peak is slightly shifted to a higher value of Q sub x y, and the distribution of intensity normal to the surface (not shown) indicates a substantially increased tilt of the lipid tails. By contrast, no change in the diffraction peak is observed after addition of myoglobin when the surface layer is maintained at a constant area (after initial compression to 40 milli newtons per meter).
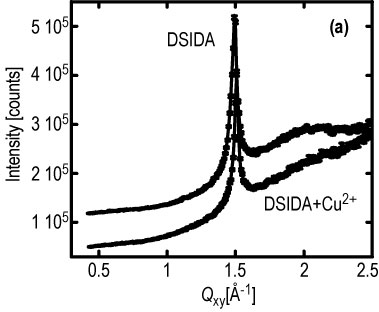
| Figure 3. Surface diffraction peak at a constant surface pressure of 40 milli newtons per meter: (a) 100 % D S I D A and also with copper 2 +, (b) 1.9 h (no peak) and 4.4 h (peak returns) after myoglobin injection. |
We hypothesize that the protein is able to insert into the lipid monolayer when it is maintained at a constant pressure of 40 milli newtons per meter, but does not insert when at constant area (surface pressure in this case rises from 40 milli newtons per meter to 43 milli newtons per meter). Thus far, all the neutron reflection data have been obtained at constant surface area. In future work we will examine adsorption to lipid monolayers composed of 100 % D S I D A at constant surface pressure. We expect to see very different arrangement of the protein adsorbed layers for conditions at which the protein inserts into the lipid monolayer versus when it does not.
In summary, a combination of neutron reflectivity and G I X D has proven to be a very powerful way to study both the protein adsorption to lipid monolayers as well as structural changes that the lipid layer itself undergoes as the protein is adsorbed. Because of different contrast conditions for neutrons and x-rays, neutron reflectivity profiles are very sensitive to the adsorption of the protein, while G I X D is ideally suited for examining in detail any changes in the 2-D ordered structure of the lipid layer itself.
This successful study opens up a rather large field to examine other important lipid-protein interactions such as insertion of biological toxins into a lipid layer. Preliminary studies of neutron reflectivity and G I X D to examine the adsorption of cholera toxin to monolayers of glycolipids (a class of lipids containing sugar molecules which are found on cell surfaces) show extraordinary promise in this regard.
References
[1] K. M. Maloney, D. R. Shnek, D. Y. Sasaki, and F. H. Arnold, Chemistry & Biology, 3, 185, (1996).
[2] M. S. Kent, H. Yim, D. Y. Sasaki, J. Majewski, G. S. Smith, K. Shin, S. K. Satija, and B. M. Ocko, Langmuir 18, 3754,(2002).
Authors
M. Kent and H. Yim
Sandia National Laboratory
Albuquerque, NM 87185
S. K. Satija
NIST Center for Neutron Research
National Institute of Standards and Technology
Gaithersburg, MD 20899-8562
J. Majewski
Los Alamos National Laboratory
Los Alamos, NM 87545
Back to FY2002 HTML main page
Go to next article
To view all symbols correctly, please download Internet Explorer 6 or Netscape 7.1